Table of Contents
Introduction
Hey everyone, data enthusiasts! Welcome to another thrilling exploration into the realm of machine learning.
Also, check Machine Learning projects:
- Machine Learning Project 1: Honda Motor Stocks best Prices analysis
- Machine Learning Project 2: Diversity in Tech Companies Best EDA Analysis
- Machine Learning Project 3: Exploring Indian Cuisine Best Analysis
- Machine Learning Project 4: Exploring Video Game Data
- Machine Learning Project 5: Best Students Performance EDA
- Machine Learning Project 6: Obesity type Best EDA and classification
- Machine Learning Project 7: Best ChatGPT Reviews Analysis
Today, we’re delving into a critical health issue that impacts millions globally – anemia. And not just any anemia, but the various types of it.
In this project, we’ll be rolling up our sleeves and diving into top-notch Exploratory Data Analysis (EDA) and classification techniques.
project machine learning
machine learning certification
certification machine learning
This project is ideal for those seeking machine learning projects for their final year or for students aiming to make a tangible impact in the real world.
Additionally, we’ll be sharing some fantastic resources and code snippets related to machine learning projects on GitHub, so you can follow along and perhaps even contribute your own enhancements.
Anemia Types EDA
import numpy as np # linear algebra import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv) import warnings warnings.filterwarnings("ignore") import matplotlib.pyplot as plt import seaborn as sns from IPython.core.display import display, HTML from sklearn.preprocessing import LabelEncoder, MinMaxScaler from imblearn.over_sampling import SMOTE from sklearn.model_selection import train_test_split from sklearn.ensemble import RandomForestClassifier from sklearn.ensemble import AdaBoostClassifier from sklearn.ensemble import ExtraTreesClassifier from sklearn.ensemble import GradientBoostingClassifier from sklearn.naive_bayes import MultinomialNB from xgboost import XGBClassifier from lightgbm import LGBMClassifier from tensorflow import keras from sklearn.metrics import accuracy_score, classification_report, confusion_matrix from collections import Counter
Exploring data
df = pd.read_csv("/kaggle/input/anemia-types-classification/diagnosed_cbc_data_v4.csv")
df.shape
(1281, 15)
df.head()
| WBC | LYMp | NEUTp | LYMn | NEUTn | RBC | HGB | HCT | MCV | MCH | MCHC | PLT | PDW | PCT | Diagnosis | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 10.0 | 43.2 | 50.1 | 4.3 | 5.0 | 2.77 | 7.3 | 24.2 | 87.7 | 26.3 | 30.1 | 189.0 | 12.5 | 0.17 | Normocytic hypochromic anemia |
| 1 | 10.0 | 42.4 | 52.3 | 4.2 | 5.3 | 2.84 | 7.3 | 25.0 | 88.2 | 25.7 | 20.2 | 180.0 | 12.5 | 0.16 | Normocytic hypochromic anemia |
| 2 | 7.2 | 30.7 | 60.7 | 2.2 | 4.4 | 3.97 | 9.0 | 30.5 | 77.0 | 22.6 | 29.5 | 148.0 | 14.3 | 0.14 | Iron deficiency anemia |
| 3 | 6.0 | 30.2 | 63.5 | 1.8 | 3.8 | 4.22 | 3.8 | 32.8 | 77.9 | 23.2 | 29.8 | 143.0 | 11.3 | 0.12 | Iron deficiency anemia |
| 4 | 4.2 | 39.1 | 53.7 | 1.6 | 2.3 | 3.93 | 0.4 | 316.0 | 80.6 | 23.9 | 29.7 | 236.0 | 12.8 | 0.22 | Normocytic hypochromic anemia |
df.isna().sum()
WBC 0 LYMp 0 NEUTp 0 LYMn 0 NEUTn 0 RBC 0 HGB 0 HCT 0 MCV 0 MCH 0 MCHC 0 PLT 0 PDW 0 PCT 0 Diagnosis 0 dtype: int64
df.describe()
| WBC | LYMp | NEUTp | LYMn | NEUTn | RBC | HGB | HCT | MCV | MCH | MCHC | PLT | PDW | PCT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| count | 1281.000000 | 1281.000000 | 1281.000000 | 1281.000000 | 1281.000000 | 1281.000000 | 1281.000000 | 1281.0000 | 1281.000000 | 1281.000000 | 1281.000000 | 1281.000000 | 1281.000000 | 1281.000000 |
| mean | 7.862717 | 25.845000 | 77.511000 | 1.880760 | 5.140940 | 4.708267 | 12.184551 | 46.1526 | 85.793919 | 32.084840 | 31.739149 | 229.981421 | 14.312512 | 0.260280 |
| std | 3.564466 | 7.038728 | 147.746273 | 1.335689 | 2.872294 | 2.817200 | 3.812897 | 104.8861 | 27.177663 | 111.170756 | 3.300352 | 93.019336 | 3.005079 | 0.685351 |
| min | 0.800000 | 6.200000 | 0.700000 | 0.200000 | 0.500000 | 1.360000 | -10.000000 | 2.0000 | -79.300000 | 10.900000 | 11.500000 | 10.000000 | 8.400000 | 0.010000 |
| 25% | 6.000000 | 25.845000 | 71.100000 | 1.880760 | 5.100000 | 4.190000 | 10.800000 | 39.2000 | 81.200000 | 25.500000 | 30.600000 | 157.000000 | 13.300000 | 0.170000 |
| 50% | 7.400000 | 25.845000 | 77.511000 | 1.880760 | 5.140940 | 4.600000 | 12.300000 | 46.1526 | 86.600000 | 27.800000 | 32.000000 | 213.000000 | 14.312512 | 0.260280 |
| 75% | 8.680000 | 25.845000 | 77.511000 | 1.880760 | 5.140940 | 5.100000 | 13.500000 | 46.1526 | 90.200000 | 29.600000 | 32.900000 | 293.000000 | 14.700000 | 0.260280 |
| max | 45.700000 | 91.400000 | 5317.000000 | 41.800000 | 79.000000 | 90.800000 | 87.100000 | 3715.0000 | 990.000000 | 3117.000000 | 92.800000 | 660.000000 | 97.000000 | 13.600000 |
Diagnoses count
fig, axes = plt.subplots(nrows=1, ncols=2, figsize=(16, 7)) counts = df["Diagnosis"].value_counts() counts.plot(kind="bar", ax=axes[0]) for container in axes[0].containers: axes[0].bar_label(container) axes[1].pie(counts, autopct="%0.2f%%", labels=counts.index) plt.tight_layout() plt.show()

machine learning projects github
machine learning projects for final year
machine learning projects for students
Correlation coefficient of patients features
corr = df[df.columns[:-1]].corr() corr.style.background_gradient(cmap='coolwarm')
| WBC | LYMp | NEUTp | LYMn | NEUTn | RBC | HGB | HCT | MCV | MCH | MCHC | PLT | PDW | PCT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WBC | 1.000000 | -0.098685 | -0.022998 | 0.379974 | 0.306216 | 0.029473 | 0.053450 | -0.002289 | -0.011190 | -0.016255 | -0.060900 | 0.211354 | -0.026318 | -0.004214 |
| LYMp | -0.098685 | 1.000000 | 0.009017 | 0.465271 | -0.296009 | -0.031355 | -0.012136 | 0.019576 | -0.015431 | -0.005443 | -0.064747 | 0.047673 | 0.011715 | -0.038542 |
| NEUTp | -0.022998 | 0.009017 | 1.000000 | -0.019970 | -0.015514 | -0.015369 | -0.025144 | -0.004367 | 0.000013 | -0.003420 | -0.004346 | 0.016326 | -0.008418 | -0.000491 |
| LYMn | 0.379974 | 0.465271 | -0.019970 | 1.000000 | 0.047662 | 0.013261 | 0.016153 | 0.003046 | -0.024092 | -0.009649 | -0.067572 | 0.072622 | -0.014422 | -0.019014 |
| NEUTn | 0.306216 | -0.296009 | -0.015514 | 0.047662 | 1.000000 | 0.029271 | 0.081665 | 0.091813 | -0.014108 | 0.047501 | -0.015920 | 0.022225 | -0.038612 | -0.043049 |
| RBC | 0.029473 | -0.031355 | -0.015369 | 0.013261 | 0.029271 | 1.000000 | 0.463446 | -0.000816 | -0.039550 | -0.009109 | -0.100370 | 0.004301 | 0.037160 | -0.013501 |
| HGB | 0.053450 | -0.012136 | -0.025144 | 0.016153 | 0.081665 | 0.463446 | 1.000000 | -0.000459 | 0.023494 | 0.001506 | -0.029115 | 0.040231 | 0.135668 | -0.053314 |
| HCT | -0.002289 | 0.019576 | -0.004367 | 0.003046 | 0.091813 | -0.000816 | -0.000459 | 1.000000 | 0.000813 | 0.608017 | 0.002065 | -0.017894 | 0.088878 | -0.012773 |
| MCV | -0.011190 | -0.015431 | 0.000013 | -0.024092 | -0.014108 | -0.039550 | 0.023494 | 0.000813 | 1.000000 | 0.013114 | 0.094950 | 0.064139 | 0.021137 | -0.043245 |
| MCH | -0.016255 | -0.005443 | -0.003420 | -0.009649 | 0.047501 | -0.009109 | 0.001506 | 0.608017 | 0.013114 | 1.000000 | 0.015006 | -0.032141 | 0.053705 | 0.008228 |
| MCHC | -0.060900 | -0.064747 | -0.004346 | -0.067572 | -0.015920 | -0.100370 | -0.029115 | 0.002065 | 0.094950 | 0.015006 | 1.000000 | 0.062829 | 0.004920 | -0.036572 |
| PLT | 0.211354 | 0.047673 | 0.016326 | 0.072622 | 0.022225 | 0.004301 | 0.040231 | -0.017894 | 0.064139 | -0.032141 | 0.062829 | 1.000000 | 0.087677 | -0.059280 |
| PDW | -0.026318 | 0.011715 | -0.008418 | -0.014422 | -0.038612 | 0.037160 | 0.135668 | 0.088878 | 0.021137 | 0.053705 | 0.004920 | 0.087677 | 1.000000 | -0.067446 |
| PCT | -0.004214 | -0.038542 | -0.000491 | -0.019014 | -0.043049 | -0.013501 | -0.053314 | -0.012773 | -0.043245 | 0.008228 | -0.036572 | -0.059280 | -0.067446 | 1.000000 |
Mean numerical values for each numerical features showin tendencies among diagnoses
index = 0 grouped = df.groupby("Diagnosis") for i in range(2): fig, axes = plt.subplots(nrows=1, ncols=7, figsize=(15, 7)) for j in range(7): mean = grouped[df.columns[index]].mean() sns.barplot(x=mean.index, y=mean, ax=axes[j]) for container in axes[j].containers: axes[j].bar_label(container, label_type="center", rotation=90) axes[j].set_xticklabels(axes[j].get_xticklabels(), rotation=90) index += 1 plt.tight_layout() plt.show()


General numerical density distribution
index = 0 for i in range(2): fig, axes = plt.subplots(nrows=1, ncols=7, figsize=(15, 6)) for j in range(7): sns.kdeplot(df, x=df.columns[index], ax=axes[j]) index += 1 plt.tight_layout() plt.show()


Density distribution among diagnoses
index = 0 for i in range(2): fig, axes = plt.subplots(nrows=1, ncols=7, figsize=(20, 6)) for j in range(7): sns.kdeplot(df, x=df.columns[index], hue="Diagnosis", ax=axes[j]) index += 1 plt.tight_layout() plt.show()


ml process
kaggle machine learning projects
Data distribution with outliers shown on boxplots
index = 0 for i in range(2): fig, axes = plt.subplots(nrows=1, ncols=7, figsize=(15, 6)) for j in range(7): sns.boxplot(df, x=df.columns[index], ax=axes[j]) index += 1 plt.tight_layout() plt.show()


Boxplots with categorical diagnostic distribution
index = 0 for i in range(2): fig, axes = plt.subplots(nrows=1, ncols=7, figsize=(15, 6)) for j in range(7): sns.boxplot(df, x="Diagnosis",y=df.columns[index], ax=axes[j]) axes[j].set_xticklabels(axes[j].get_xticklabels(), rotation=90) index += 1 plt.tight_layout() plt.show()


Encoding diagnoses
le = LabelEncoder() df["Diagnosis"] = le.fit_transform(df["Diagnosis"].values)
x = df.iloc[:, :-1].values y = df.iloc[:, -1].values
Scaling features
scaler = MinMaxScaler() x = scaler.fit_transform(x)
x_train, x_test, y_train, y_test = train_test_split(x, y, random_state=42, test_size=0.2)
Handling class imbalances applying SMOTE
smote = SMOTE() print("Before: ", Counter(y_train)) x_train, y_train = smote.fit_resample(x_train, y_train) print("After: ", Counter(y_train))
Before: Counter({0: 255, 5: 223, 6: 223, 1: 155, 8: 56, 7: 45, 2: 42, 4: 17, 3: 8})
After: Counter({5: 255, 6: 255, 0: 255, 8: 255, 7: 255, 1: 255, 2: 255, 4: 255, 3: 255})
step machine learning
step of machine learning
ml projects
Machine Learning Models
rfc = RandomForestClassifier() abc = AdaBoostClassifier() etc = ExtraTreesClassifier() gbc = GradientBoostingClassifier() mnb = MultinomialNB() xgb = XGBClassifier() lgb = LGBMClassifier() models = [rfc, abc, etc, gbc, mnb, xgb, lgb] names = ["Random Forest", "Ada Boost", "Extra Trees", "Gradient Boosting", "Nave Bayes", "XGBoost", "LightGBM"]
Neural Network architecture
model = keras.models.Sequential([ keras.layers.Dense(32, activation="relu"), keras.layers.Dropout(0.2), keras.layers.Dense(df["Diagnosis"].nunique(), activation="softmax") ]) model.compile(optimizer='adam', loss="categorical_crossentropy", metrics=['accuracy']) model.summary()
Model: "sequential"
┏━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━┓ ┃ Layer (type) ┃ Output Shape ┃ Param # ┃ ┡━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━┩ │ dense (Dense) │ ? │ 0 (unbuilt) │ ├─────────────────────────────────┼────────────────────────┼───────────────┤ │ dropout (Dropout) │ ? │ 0 │ ├─────────────────────────────────┼────────────────────────┼───────────────┤ │ dense_1 (Dense) │ ? │ 0 (unbuilt) │ └─────────────────────────────────┴────────────────────────┴───────────────┘
Total params: 0 (0.00 B)
Trainable params: 0 (0.00 B)
Non-trainable params: 0 (0.00 B)
Training models
history = model.fit(x_train, y_cat, validation_split=0.1, epochs=50, batch_size=32)
ANN model performance during training – history log
fig, axes = plt.subplots(nrows=1, ncols=2, figsize=(10, 7)) axes[0].plot(history.history["loss"], label="Training loss") axes[0].plot(history.history["val_loss"], label="Validation loss") axes[0].legend() axes[0].set_title("Loss log") axes[1].plot(history.history["accuracy"], label="Training accuracy") axes[1].plot(history.history["val_accuracy"], label="Validation accuracy") axes[1].legend() axes[1].set_title("Accuracy log") plt.show()

ml project
machine learning python projects
machine learning projects in python
Training ML models
scores, reports, cms = [], dict(), dict() for m, n in zip(models, names): score, report, cm = training(m) scores += [score*100] reports[n] = report cms[n] = cm pred = model.predict(x_test) pred = [np.argmax(i) for i in pred] scores += [accuracy_score(pred, y_test)*100] reports["ANN"] = classification_report(pred, y_test) cms["ANN"] = confusion_matrix(pred, y_test) names += ["ANN"]
ML models accuracies from worst to best
dt = pd.DataFrame({"scores": scores}, index=names) dt = dt.sort_values("scores", ascending=False) dt["scores"] = round(dt["scores"], 2) fig, axes = plt.subplots() sns.barplot(x=dt.index, y=dt.iloc[:, 0], ax=axes) for container in axes.containers: axes.bar_label(container) axes.set_xticklabels(axes.get_xticklabels(), rotation=90) plt.show()

Confusion matrices from best to worst performing model
index = 0 for i in range(2): fig, axes = plt.subplots(nrows=1, ncols=4, figsize=(10, 7)) for j in range(4): sns.heatmap(cms[dt.index[index]], annot=True, ax=axes[j]) axes[j].set_title("{}: {}%".format(dt.index[index], dt.iloc[index, 0])) index += 1 plt.tight_layout() plt.show()


Classification reports
for i in dt.index: print("*"*30) print(i) print(reports[i]) print("\n")
******************************
LightGBM
precision recall f1-score support
0 1.00 1.00 1.00 81
1 1.00 1.00 1.00 34
2 1.00 1.00 1.00 5
3 1.00 0.75 0.86 4
4 1.00 1.00 1.00 1
5 1.00 1.00 1.00 56
6 0.98 1.00 0.99 45
7 1.00 1.00 1.00 14
8 1.00 1.00 1.00 17
accuracy 1.00 257
macro avg 1.00 0.97 0.98 257
weighted avg 1.00 1.00 1.00 257
******************************
Random Forest
precision recall f1-score support
0 0.99 1.00 0.99 80
1 1.00 1.00 1.00 34
2 1.00 0.83 0.91 6
3 1.00 1.00 1.00 3
4 1.00 0.50 0.67 2
5 1.00 1.00 1.00 56
6 0.98 1.00 0.99 45
7 1.00 1.00 1.00 14
8 1.00 1.00 1.00 17
accuracy 0.99 257
macro avg 1.00 0.93 0.95 257
weighted avg 0.99 0.99 0.99 257
******************************
XGBoost
precision recall f1-score support
0 0.99 1.00 0.99 80
1 1.00 1.00 1.00 34
2 1.00 1.00 1.00 5
3 1.00 0.75 0.86 4
4 1.00 1.00 1.00 1
5 1.00 1.00 1.00 56
6 0.98 1.00 0.99 45
7 1.00 0.93 0.97 15
8 1.00 1.00 1.00 17
accuracy 0.99 257
macro avg 1.00 0.96 0.98 257
weighted avg 0.99 0.99 0.99 257
******************************
Gradient Boosting
precision recall f1-score support
0 0.99 1.00 0.99 80
1 0.97 1.00 0.99 33
2 1.00 1.00 1.00 5
3 1.00 0.75 0.86 4
4 1.00 1.00 1.00 1
5 1.00 1.00 1.00 56
6 0.98 1.00 0.99 45
7 1.00 0.88 0.93 16
8 1.00 1.00 1.00 17
accuracy 0.99 257
macro avg 0.99 0.96 0.97 257
weighted avg 0.99 0.99 0.99 257
******************************
Extra Trees
precision recall f1-score support
0 0.91 0.95 0.93 78
1 0.91 0.91 0.91 34
2 0.60 0.60 0.60 5
3 0.67 1.00 0.80 2
4 0.00 0.00 0.00 3
5 0.91 0.81 0.86 63
6 0.76 0.97 0.85 36
7 0.93 0.93 0.93 14
8 0.94 0.73 0.82 22
accuracy 0.88 257
macro avg 0.74 0.77 0.74 257
weighted avg 0.88 0.88 0.87 257
******************************
Ada Boost
precision recall f1-score support
0 1.00 0.75 0.86 108
1 0.00 0.00 0.00 0
2 0.00 0.00 0.00 0
3 0.00 0.00 0.00 0
4 0.00 0.00 0.00 0
5 0.00 0.00 0.00 0
6 0.98 0.30 0.46 149
7 0.00 0.00 0.00 0
8 0.00 0.00 0.00 0
accuracy 0.49 257
macro avg 0.22 0.12 0.15 257
weighted avg 0.99 0.49 0.63 257
******************************
ANN
precision recall f1-score support
0 0.69 0.71 0.70 79
1 0.44 0.60 0.51 25
2 1.00 0.36 0.53 14
3 1.00 0.20 0.33 15
4 1.00 0.02 0.04 52
5 0.21 0.38 0.27 32
6 0.22 0.48 0.30 21
7 0.00 0.00 0.00 18
8 0.00 0.00 0.00 1
accuracy 0.40 257
macro avg 0.51 0.30 0.30 257
weighted avg 0.62 0.40 0.38 257
******************************
Nave Bayes
precision recall f1-score support
0 0.68 0.63 0.65 87
1 0.26 0.35 0.30 26
2 0.60 0.25 0.35 12
3 1.00 0.25 0.40 12
4 1.00 0.04 0.07 27
5 0.05 0.19 0.08 16
6 0.17 0.40 0.24 20
7 0.00 0.00 0.00 0
8 0.94 0.28 0.43 57
accuracy 0.38 257
macro avg 0.52 0.26 0.28 257
weighted avg 0.66 0.38 0.41 257
ml projects ideas
project manager artificial intelligence
best machine learning courses reddit
machine learning projects for resume
Anemia Types Using Ensemble Learning
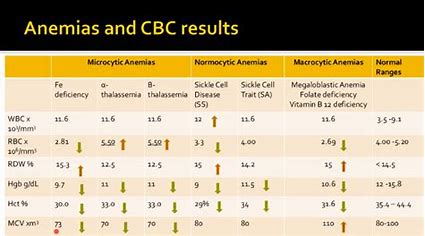
About dataset
- About Dataset
- CBC data labeled with the diagnosis of Anemia type, The data collected among several CBCs data and diagnosed manually
- Data Dictionary:
- HGB: The amount of hemoglobin in the blood, crucial for oxygen transport.
- PlT: The number of platelets in the blood, involved in blood clotting.
- WBC: The count of white blood cells, vital for immune response.
- RBC: The count of red blood cells, responsible for oxygen transport.
- MCV (Mean Corpuscular Volume): Average volume of a single red blood cell.
- MCH (Mean Corpuscular Hemoglobin): Average amount of hemoglobin per red blood cell.
- MCHC (Mean Corpuscular Hemoglobin Concentration): Average concentration of hemoglobin in red blood cells.
- PDW: a measurement of the variability in platelet size distribution in the blood
- PCT: A procalcitonin test can help your health care provider diagnose if you have sepsis from a bacterial infection or if you have a high risk of developing sepsis
- Diagnosis: Anemia type based on the CBC parameters
import pandas as pd import matplotlib.pyplot as plt import seaborn as sns import numpy as np import warnings
df = pd.read_csv('/kaggle/input/anemia-types-classification/diagnosed_cbc_data_v4.csv')
df.sample(10)
| WBC | LYMp | NEUTp | LYMn | NEUTn | RBC | HGB | HCT | MCV | MCH | MCHC | PLT | PDW | PCT | Diagnosis | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 694 | 8.70 | 25.845 | 77.511 | 1.88076 | 5.14094 | 5.70 | 14.5 | 46.1526 | 95.0 | 31.0 | 33.0 | 380.0 | 14.700000 | 0.26028 | Healthy |
| 1077 | 4.30 | 25.845 | 77.511 | 1.88076 | 5.14094 | 2.71 | 8.0 | 46.1526 | 87.8 | 29.5 | 33.6 | 32.0 | 14.312512 | 0.26028 | Normocytic normochromic anemia |
| 445 | 8.50 | 27.800 | 62.400 | 2.40000 | 5.30000 | 3.85 | 11.2 | 34.8000 | 90.6 | 29.0 | 32.1 | 243.0 | 13.800000 | 0.23000 | Normocytic normochromic anemia |
| 339 | 7.40 | 25.900 | 67.100 | 1.90000 | 5.00000 | 5.29 | 10.3 | 34.8000 | 65.8 | 19.4 | 29.5 | 37.0 | 8.400000 | 0.03000 | Iron deficiency anemia |
| 719 | 8.70 | 25.845 | 77.511 | 1.88076 | 5.14094 | 5.70 | 14.5 | 46.1526 | 95.0 | 31.0 | 33.0 | 380.0 | 13.900000 | 0.26028 | Healthy |
| 730 | 7.20 | 25.845 | 77.511 | 1.88076 | 5.14094 | 5.50 | 13.5 | 46.1526 | 93.0 | 31.0 | 32.0 | 330.0 | 18.200000 | 0.26028 | Healthy |
| 438 | 4.20 | 9.500 | 84.700 | 0.40000 | 3.60000 | 3.34 | 9.2 | 28.4000 | 85.3 | 275.0 | 32.3 | 107.0 | 13.100000 | 0.10000 | Normocytic normochromic anemia |
| 770 | 8.20 | 25.845 | 77.511 | 1.88076 | 5.14094 | 5.30 | 14.8 | 46.1526 | 91.0 | 30.0 | 33.0 | 360.0 | 14.200000 | 0.26028 | Healthy |
| 320 | 6.20 | 16.300 | 77.200 | 1.00000 | 4.80000 | 3.85 | 9.3 | 31.5000 | 82.0 | 24.1 | 29.5 | 161.0 | 12.300000 | 0.15000 | Normocytic hypochromic anemia |
| 997 | 4.48 | 25.845 | 77.511 | 1.88076 | 5.14094 | 4.87 | 11.9 | 46.1526 | 82.5 | 24.4 | 29.6 | 384.0 | 14.312512 | 0.26028 | Normocytic hypochromic anemia |
df.shape
(1281, 15)
df.info()
<class 'pandas.core.frame.DataFrame'> RangeIndex: 1281 entries, 0 to 1280 Data columns (total 15 columns): # Column Non-Null Count Dtype --- ------ -------------- ----- 0 WBC 1281 non-null float64 1 LYMp 1281 non-null float64 2 NEUTp 1281 non-null float64 3 LYMn 1281 non-null float64 4 NEUTn 1281 non-null float64 5 RBC 1281 non-null float64 6 HGB 1281 non-null float64 7 HCT 1281 non-null float64 8 MCV 1281 non-null float64 9 MCH 1281 non-null float64 10 MCHC 1281 non-null float64 11 PLT 1281 non-null float64 12 PDW 1281 non-null float64 13 PCT 1281 non-null float64 14 Diagnosis 1281 non-null object dtypes: float64(14), object(1) memory usage: 150.2+ KB
df.describe()
| WBC | LYMp | NEUTp | LYMn | NEUTn | RBC | HGB | HCT | MCV | MCH | MCHC | PLT | PDW | PCT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| count | 1281.000000 | 1281.000000 | 1281.000000 | 1281.000000 | 1281.000000 | 1281.000000 | 1281.000000 | 1281.0000 | 1281.000000 | 1281.000000 | 1281.000000 | 1281.000000 | 1281.000000 | 1281.000000 |
| mean | 7.862717 | 25.845000 | 77.511000 | 1.880760 | 5.140940 | 4.708267 | 12.184551 | 46.1526 | 85.793919 | 32.084840 | 31.739149 | 229.981421 | 14.312512 | 0.260280 |
| std | 3.564466 | 7.038728 | 147.746273 | 1.335689 | 2.872294 | 2.817200 | 3.812897 | 104.8861 | 27.177663 | 111.170756 | 3.300352 | 93.019336 | 3.005079 | 0.685351 |
| min | 0.800000 | 6.200000 | 0.700000 | 0.200000 | 0.500000 | 1.360000 | -10.000000 | 2.0000 | -79.300000 | 10.900000 | 11.500000 | 10.000000 | 8.400000 | 0.010000 |
| 25% | 6.000000 | 25.845000 | 71.100000 | 1.880760 | 5.100000 | 4.190000 | 10.800000 | 39.2000 | 81.200000 | 25.500000 | 30.600000 | 157.000000 | 13.300000 | 0.170000 |
| 50% | 7.400000 | 25.845000 | 77.511000 | 1.880760 | 5.140940 | 4.600000 | 12.300000 | 46.1526 | 86.600000 | 27.800000 | 32.000000 | 213.000000 | 14.312512 | 0.260280 |
| 75% | 8.680000 | 25.845000 | 77.511000 | 1.880760 | 5.140940 | 5.100000 | 13.500000 | 46.1526 | 90.200000 | 29.600000 | 32.900000 | 293.000000 | 14.700000 | 0.260280 |
| max | 45.700000 | 91.400000 | 5317.000000 | 41.800000 | 79.000000 | 90.800000 | 87.100000 | 3715.0000 | 990.000000 | 3117.000000 | 92.800000 | 660.000000 | 97.000000 | 13.600000 |
df.dtypes
WBC float64 LYMp float64 NEUTp float64 LYMn float64 NEUTn float64 RBC float64 HGB float64 HCT float64 MCV float64 MCH float64 MCHC float64 PLT float64 PDW float64 PCT float64 Diagnosis object dtype: object
df.isnull().sum()
WBC 0 LYMp 0 NEUTp 0 LYMn 0 NEUTn 0 RBC 0 HGB 0 HCT 0 MCV 0 MCH 0 MCHC 0 PLT 0 PDW 0 PCT 0 Diagnosis 0 dtype: int64
machine learning project for resume
best machine learning projects
cool machine learning projects
data visulization
plt.figure(figsize=(10, 6)) sns.countplot(x='Diagnosis', data=df) plt.title('Count of Each Diagnosis Category') plt.xlabel('Diagnosis') plt.ylabel('Count') plt.xticks(rotation=90) plt.show()

columns = ['WBC', 'LYMp', 'NEUTp', 'LYMn', 'NEUTn', 'RBC', 'HGB', 'HCT', 'MCV', 'MCH', 'MCHC', 'PLT', 'PDW', 'PCT'] for column in columns: plt.figure(figsize=(8, 5)) sns.histplot(df[column], kde=True) plt.title(f'Distribution of {column}') plt.xlabel(column) plt.ylabel('Frequency') plt.xticks(rotation=90) plt.show()
/opt/conda/lib/python3.10/site-packages/seaborn/_oldcore.py:1119: FutureWarning: use_inf_as_na option is deprecated and will be removed in a future version. Convert inf values to NaN before operating instead.
with pd.option_context('mode.use_inf_as_na', True):

/opt/conda/lib/python3.10/site-packages/seaborn/_oldcore.py:1119: FutureWarning: use_inf_as_na option is deprecated and will be removed in a future version. Convert inf values to NaN before operating instead.
with pd.option_context('mode.use_inf_as_na', True):

/opt/conda/lib/python3.10/site-packages/seaborn/_oldcore.py:1119: FutureWarning: use_inf_as_na option is deprecated and will be removed in a future version. Convert inf values to NaN before operating instead.
with pd.option_context('mode.use_inf_as_na', True):

/opt/conda/lib/python3.10/site-packages/seaborn/_oldcore.py:1119: FutureWarning: use_inf_as_na option is deprecated and will be removed in a future version. Convert inf values to NaN before operating instead.
with pd.option_context('mode.use_inf_as_na', True):

/opt/conda/lib/python3.10/site-packages/seaborn/_oldcore.py:1119: FutureWarning: use_inf_as_na option is deprecated and will be removed in a future version. Convert inf values to NaN before operating instead.
with pd.option_context('mode.use_inf_as_na', True):

/opt/conda/lib/python3.10/site-packages/seaborn/_oldcore.py:1119: FutureWarning: use_inf_as_na option is deprecated and will be removed in a future version. Convert inf values to NaN before operating instead.
with pd.option_context('mode.use_inf_as_na', True):

/opt/conda/lib/python3.10/site-packages/seaborn/_oldcore.py:1119: FutureWarning: use_inf_as_na option is deprecated and will be removed in a future version. Convert inf values to NaN before operating instead.
with pd.option_context('mode.use_inf_as_na', True):

/opt/conda/lib/python3.10/site-packages/seaborn/_oldcore.py:1119: FutureWarning: use_inf_as_na option is deprecated and will be removed in a future version. Convert inf values to NaN before operating instead.
with pd.option_context('mode.use_inf_as_na', True):

/opt/conda/lib/python3.10/site-packages/seaborn/_oldcore.py:1119: FutureWarning: use_inf_as_na option is deprecated and will be removed in a future version. Convert inf values to NaN before operating instead.
with pd.option_context('mode.use_inf_as_na', True):
step machine learning
step of machine learning
ml projects
ml project

/opt/conda/lib/python3.10/site-packages/seaborn/_oldcore.py:1119: FutureWarning: use_inf_as_na option is deprecated and will be removed in a future version. Convert inf values to NaN before operating instead.
with pd.option_context('mode.use_inf_as_na', True):

/opt/conda/lib/python3.10/site-packages/seaborn/_oldcore.py:1119: FutureWarning: use_inf_as_na option is deprecated and will be removed in a future version. Convert inf values to NaN before operating instead.
with pd.option_context('mode.use_inf_as_na', True):

/opt/conda/lib/python3.10/site-packages/seaborn/_oldcore.py:1119: FutureWarning: use_inf_as_na option is deprecated and will be removed in a future version. Convert inf values to NaN before operating instead.
with pd.option_context('mode.use_inf_as_na', True):

/opt/conda/lib/python3.10/site-packages/seaborn/_oldcore.py:1119: FutureWarning: use_inf_as_na option is deprecated and will be removed in a future version. Convert inf values to NaN before operating instead.
with pd.option_context('mode.use_inf_as_na', True):

/opt/conda/lib/python3.10/site-packages/seaborn/_oldcore.py:1119: FutureWarning: use_inf_as_na option is deprecated and will be removed in a future version. Convert inf values to NaN before operating instead.
with pd.option_context('mode.use_inf_as_na', True):

import seaborn as sns import matplotlib.pyplot as plt # List of columns to plot columns = ['WBC', 'LYMp', 'NEUTp', 'LYMn', 'NEUTn', 'RBC', 'HGB', 'HCT', 'MCV', 'MCH', 'MCHC', 'PLT', 'PDW', 'PCT'] # Loop through each column and create a boxplot for column in columns: plt.figure(figsize=(8, 5)) sns.boxplot(x=df[column]) # Correct usage of boxplot without kde plt.title(f'Distribution of {column}') plt.xlabel(column) plt.show()










projects on machine learning
machine learning project




Train_test_split
x = df.drop(columns = ['Diagnosis']) y = df['Diagnosis']
from sklearn.preprocessing import LabelEncoder label_encoder = LabelEncoder() y_label = label_encoder.fit_transform(y)
y_label
array([5, 5, 1, ..., 0, 0, 0])
from sklearn.model_selection import train_test_split x_train,x_test,y_train,y_test = train_test_split(x,y_label,test_size =0.2,random_state =43)
Ensemble Learning
from sklearn.ensemble import RandomForestClassifier from sklearn.neighbors import KNeighborsClassifier from sklearn.linear_model import LogisticRegression from sklearn.ensemble import GradientBoostingClassifier
estimators = [ ('rf', RandomForestClassifier(n_estimators=10, random_state=42)), ('knn', KNeighborsClassifier(n_neighbors=10)), ('gbdt',GradientBoostingClassifier()) ]
machine learning projects github
machine learning projects for final year
machine learning projects for students
from sklearn.ensemble import StackingClassifier clf = StackingClassifier( estimators=estimators, final_estimator=LogisticRegression(), cv=10 )
clf.fit(x_train,y_train)
StackingClassifier
StackingClassifier(cv=10,
estimators=[('rf',
RandomForestClassifier(n_estimators=10,
random_state=42)),
('knn', KNeighborsClassifier(n_neighbors=10)),
('gbdt', GradientBoostingClassifier())],
final_estimator=LogisticRegression())rfRandomForestClassifier
RandomForestClassifier(n_estimators=10, random_state=42)
knnKNeighborsClassifier
KNeighborsClassifier(n_neighbors=10)
gbdtGradientBoostingClassifier
GradientBoostingClassifier()
final_estimatorLogisticRegression
LogisticRegression()
y_pred = clf.predict(x_test)
from sklearn.metrics import accuracy_score accuracy_score(y_test,y_pred)
0.980544747081712
# XGB boost
import xgboost as xgb xgb_model = xgb.XGBClassifier() xgb_model.fit(x_train, y_train) y_pred = xgb_model.predict(x_test) accuracy = accuracy_score(y_test, y_pred) print("Accuracy:", accuracy)
Accuracy: 0.980544747081712
Conclusion
Hey there, data warriors! Great news! We successfully conquered anemia classification using machine learning. We delved into the data and created intelligent models.
To all the aspiring project managers in the field of machine learning, this project highlighted the significance of having clear objectives, well-structured processes, and effective communication. It’s not only about algorithms but also about seamless coordination.
machine learning project manager
machine learning project management
machine learning projects for masters students
For those of you who are working on machine learning projects for masters students, this is just the beginning. Utilize these skills to take on even more significant challenges in the future.
Our main goal was to demystify anemia using data, and we absolutely achieved that. Keep your curiosity alive and happy coding!


0 Comments